Research
Identification and functional analysis of disease susceptibility genes for cardiovascular and metabolic diseases
Unlike congenital diseases in children, most adult-onset diseases are caused by many different genes that increase the likelihood of developing a disease, together with environmental factors such as diet and lifestyle. Such genes are called disease susceptibility genes. In 2013, a research group including us analyzed Takayasu’s arteritis, one of the most common autoimmune diseases in the cardiovascular field, using genome-wide association analysis, and reported for the first time that the gene IL12B is a disease susceptibility gene for Takayasu’s arteritis. This gene is also known as a disease susceptibility gene for other tissue autoimmune diseases, ulcerative colitis, Crohn’s disease, and psoriasis, and these diseases were revealed to be genetically related diseases (Am J Hum Genet. 2013; 93(2):289-297, Arthritis Rheumatol. 2015;67(8):2226-32, Proc Natl Acad Sci U S A. 2018;115(51):13045-13050, etc.). Ustekinumab, a drug that suppresses the action of IL12, was originally known to treat psoriasis, but then accepted as a drug for Crohn’s disease in 2017, for ulcerative colitis in 2020, and recently some reports suggest that this drug also works for Takayasu arteritis. It is a great pleasure to see the research we were involved in gradually being applied to the actual treatment of patients over the years, and we once again feel that our job is challenging and rewarding.
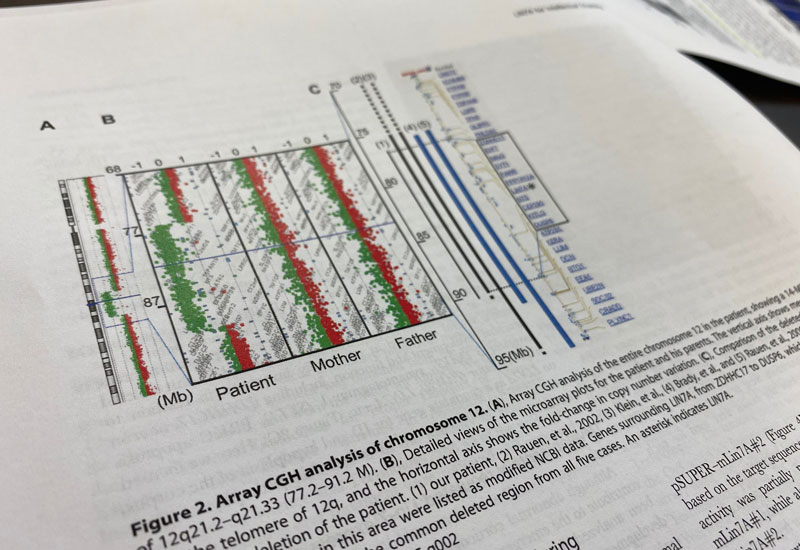
Identification of causative genes for pediatric congenital and adult-onset monogenic diseases
Our laboratory is also in charge of the Genetic Counseling Division of our hospital and handles many requests for genetic analysis in close collaboration with the Department of Pediatrics and Obstetrics & Gynecology. Led by Associate Professor Matsumoto, we have performed array CGH analyses (analysis to detect DNA copy number changes across all chromosomes) and whole exome analyses for a wide variety of diseases including autism spectrum disorders, developmental disorders, intellectual developmental disorders, and malformation syndromes.
We showed that polymorphisms of body clock-related genes are common in autism spectrum disorders with or without sleep disorders, and clarified their involvement in the pathology (Brain & development 2015; 38: 91-99). We are also actively engaged in collaborative research with other institutions. For example, the effects of a genetic variation on cortical structure and neuroaxonal elongation were studied in collaboration with the Aichi Developmental Disability Center (PloS one 2014;9: e92695, Journal of neurochemistry 2015; 132: 61-69 ). In addition, we are conducting analyses in epilepsy and cardiac diseases (Brain & development 2021; 43: 857-862, Molecular genetics & genomic medicine 2022: e2008).
We also perform genetic testing for prenatal diagnosis using amniotic fluid cells. In addition to already established methodologies for sex determination and diagnosis of monogenic disorders, we are conducting clinical research to develop new diagnostic methods, such as measuring the skewing in X-chromosome inactivation in children with X-linked diseases (Sci Rep 2024 14(1):440).
Genetic analyses of adult-onset familial diseases have also been conducted. In one familial leukemia family, we reported that a gene mutation caused the transcription factor RUNX1 to be unmethylated (Blood 2020 136(17):1919-1932). Changes in the activity of a protein by phosphorylation, acetylation, or methylation are called post-translational modifications (PTM). Although it has long been said that such PTM are important in regulating the activity of transcription factors, there are not many cases in which abnormalities in PTM of transcription factors actually lead to diseases such as leukemia. Our laboratory is also focusing its research on phenomena that have been described in textbooks but have not been properly proven.
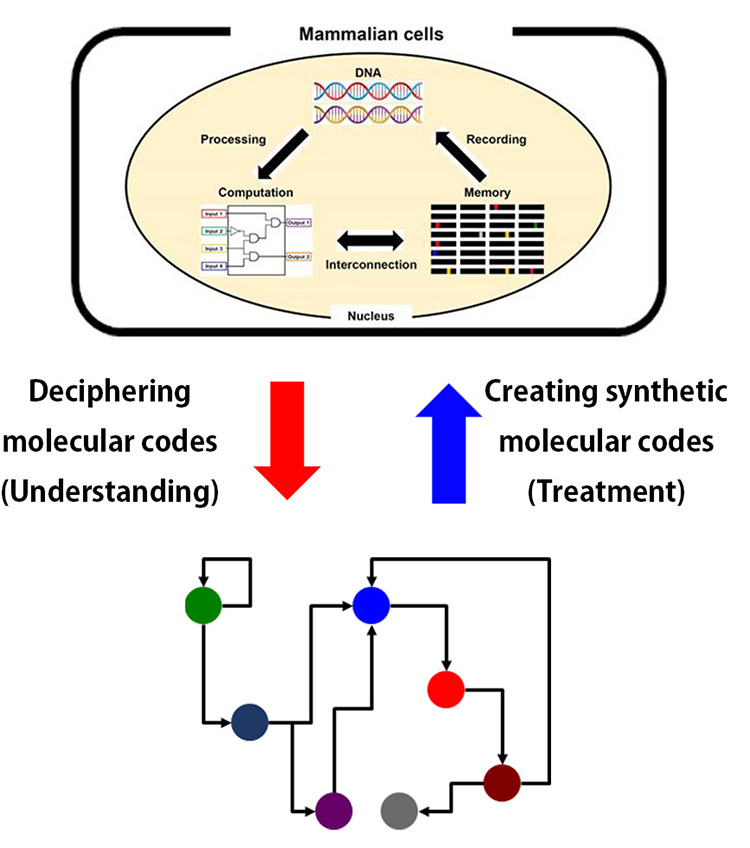
Elucidating molecular codes that cause cardiovascular disease and developing novel therapeutic platforms using synthetic gene circuits
We are developing novel therapeutic strategies that eradicate heart failure.
Heart failure is a final common pathway of various heart diseases. Pathological stress, including ischemia and pressure overload, causes enlargement of the heart and impairs its pumping function. The prevalence of heart failure is rapidly increasing in aging societies. Heart failure is associated with a decline in cognitive function and daily living activity in the elderly. The current optimal medication therapy can slow the progression of heart failure, but cannot cure heart failure. Novel therapies that can cure heart failure are urgently needed.
To address this issue, our group has been elucidating molecular mechanisms of heart failure and developing novel therapeutic platforms. Using genome-scale phenotypic screening, we have identified the essential genes that are associated with the pathogenesis of heart failure and have been analyzing their function.
To cure heart failure, it is important to induce the expression of the optimal therapeutic molecules at the optimal timing and level in the optimal location, due to its multifactorial nature. To address this challenge, we are developing artificial biological programs that control cells or molecules with spatiotemporal precision, by designing synthetic promoters, transcription factors, and receptors. Our ultimate goal is to develop therapeutic platforms that enable automatic self-restoration in the human body.
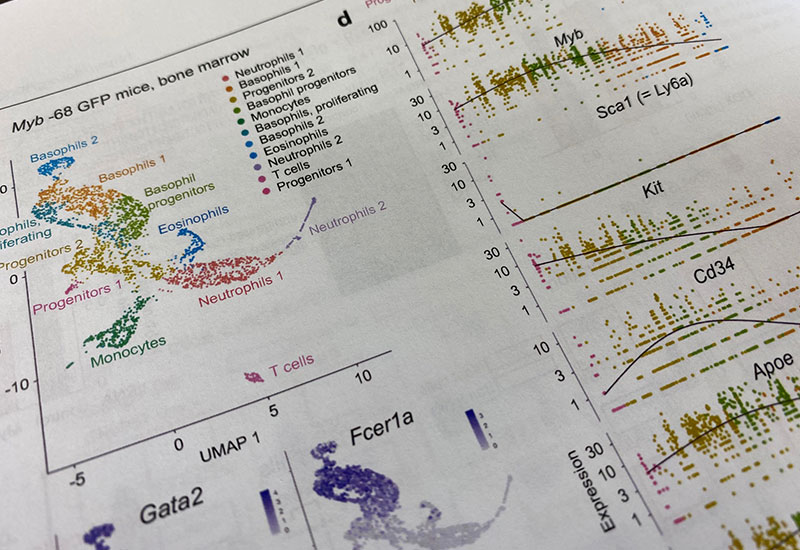
Unlocking the mysteries of life using bioinformatics
Next-generation sequencers are essential for human genetics research. For this reason, many researchers in human genetics are somewhat skilled in bioinformatics. We apply the knowledge of bioinformatics cultivated through genetic analysis to research for many other life phenomena in the human body. Our recent achievements include a comprehensive genetic analysis of basophil differentiation in bone marrow at the single-cell level (Nat Commun. 2022 13(1):7064). In this study, we succeeded in revealing the differentiation process of basophils in the bone marrow from hematopoietic stem cells to mature basophils in an uninterrupted trajectory.
The international journal Science chose single-cell analysis as the number one innovative technology of 2018. COVID-19 vaccines were chosen for 2020, so single-cell analysis is as innovative a technology as COVID-19 vaccines. However, the analysis requires special knowledge of bioinformatics. Our laboratory has installed equipment for single-cell analysis, and we respond to a wide range of requests for data analysis from inside and outside our university.
