RESEARCH
Mitochondria
1. Comprehensive analysis of novel mitochondrial membrane proteins
We have performed proteome analysis of mitochondrial membrane proteins.
To date, we have identified 171 proteins from purified mitochondrial membrane fraction of HeLa cells by 2D-electrophoresis and LC-MS/MS. Among them, we focus on 13 novel proteins, which have no information about the function and localization. Eight proteins were subcloned from them, and we confirmed that 7 proteins actually localize to the mitochondria.
We are now investigating functions of the newly identified mitochondrial membrane proteins by biochemical and molecular biological approach.
2. Fuctional analysis of mitochondrial protein, PHB
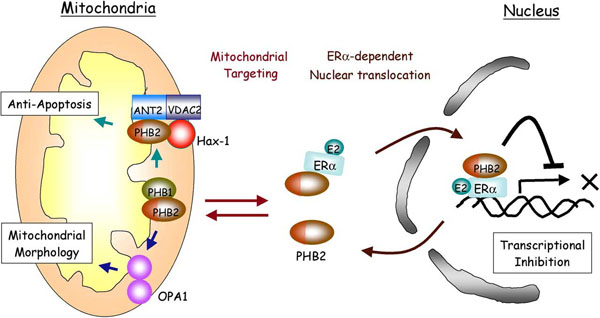
Prohibitin (PHB) are evolutionally conserved mitochondrial proteins, which have pleiotropic cellular functions, including apoptosis, regulation of cell cycle, signal transduction, and aging.
Recently, we have initially identified mitochondrial functions of human PHB2, such as anti-apoptotic function and regulation of the mitochondrial morphology (Kasashima et al, 2006). PHBs have been known to act as transcriptional repressor in the nucleus, suggesting that PHBs function in both the organelles and couple nuclear-mitochondrial interaction.
We are now investigating the novel mitochondrial functions of PHBs and the relation with their nuclear function.
Summary of the localization and functions of PHB2.
RNA Processing & Gene Regulation
Our laboratory studies the mechanisms of gene expression on the posttranscriptional level. One is the molecular details of how tissue-specific alternative splicing, i.e., inclusion or skipping of a variably spliced exon, occurs in mammalian cells.
To investigate the mechanism of alternative splicing, we work on human mitochondrial ATP synthase gamma subunit gene (F1gamma) whose alternative exon 9 is excluded only in heart and skeletal muscle as a tissue-specific model substrate.
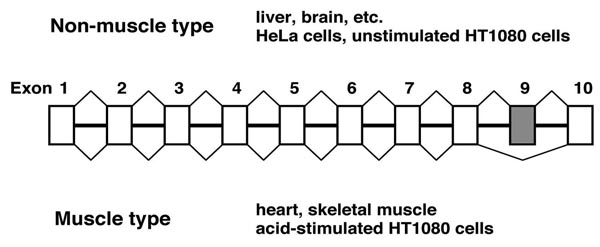
Tissue-specific alternative splicing in F1γ pre-mRNA.
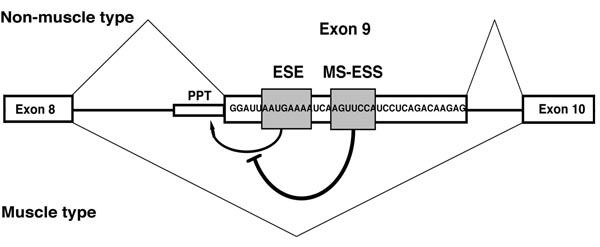
Selection of F1γ exon 9 is regulated by two cis-acting regulatory elements in the same exon.
Another area is to determine how RNA-recognition motif (RRM)-type RNA-binding proteins regulate gene expression during development and during cellular differentiation using mouse and rat models. Developmentally-regulated RNA-binding protein (Drb1) consists of four RRMs was newly cloned in our laboratory on the screening of neuron-specific RNA-binding proteins.
drb1 gene is specifically expressed in fetal rat brain and gradually reduced during development. Our current goals are to (i) identify RNA target(s); (ii) understand the mechanism of regulation; (iii) make drb1 knockout mice.
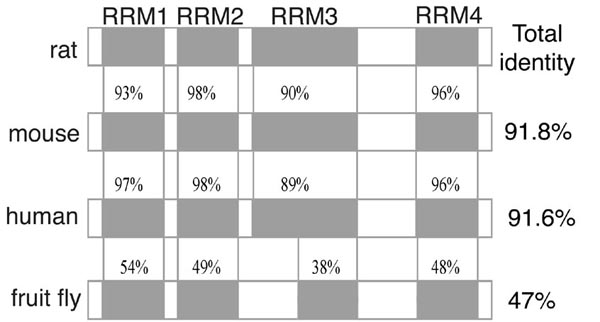
Amino acid identity between rat, mouse, human and fruit fly Drb1
We also investigate intracellular movements of splicing factors including a subset of SR protein family, in response to UV- and heat shock-induced stress signals.